
Jeff Guo
ML Research Scientist
Isomorphic Labs
I'm a research scientist at Isomorphic Labs in Lausanne, Switzerland. I completed my PhD at EPFL, advised by Philippe Schwaller, where I developed language-based generative models for synthesizable small molecule design. During my PhD, I interned at Microsoft AI4Science. Before that, I worked at AstraZeneca in the Molecular AI department on REINVENT (generative model suite) and applied it on commercial drug discovery projects.
Some research questions I'm fascinated about are:
- Given an arbitrary optimization objective, how can generative models reliably sample optimal molecules?
- How can the synthesizability of small molecules be made maximally actionable (can actually be made in the lab)?
- How can multimodal generative models improve molecular design (and be fine-tuned)?
Experience
Research Scientist — Isomorphic Labs
ML for drug discovery.
PhD Student — EPFL
Developing language-based generative models for synthesizable small molecule design.
Research Intern — Microsoft AI4Science
ML for chemistry.
Graduate Scientist — AstraZeneca
Expanded the capabilities of REINVENT (generative model suite) and applied it on commercial drug discovery projects.
Selected Publications
For up-to-date publications, see Google Scholar
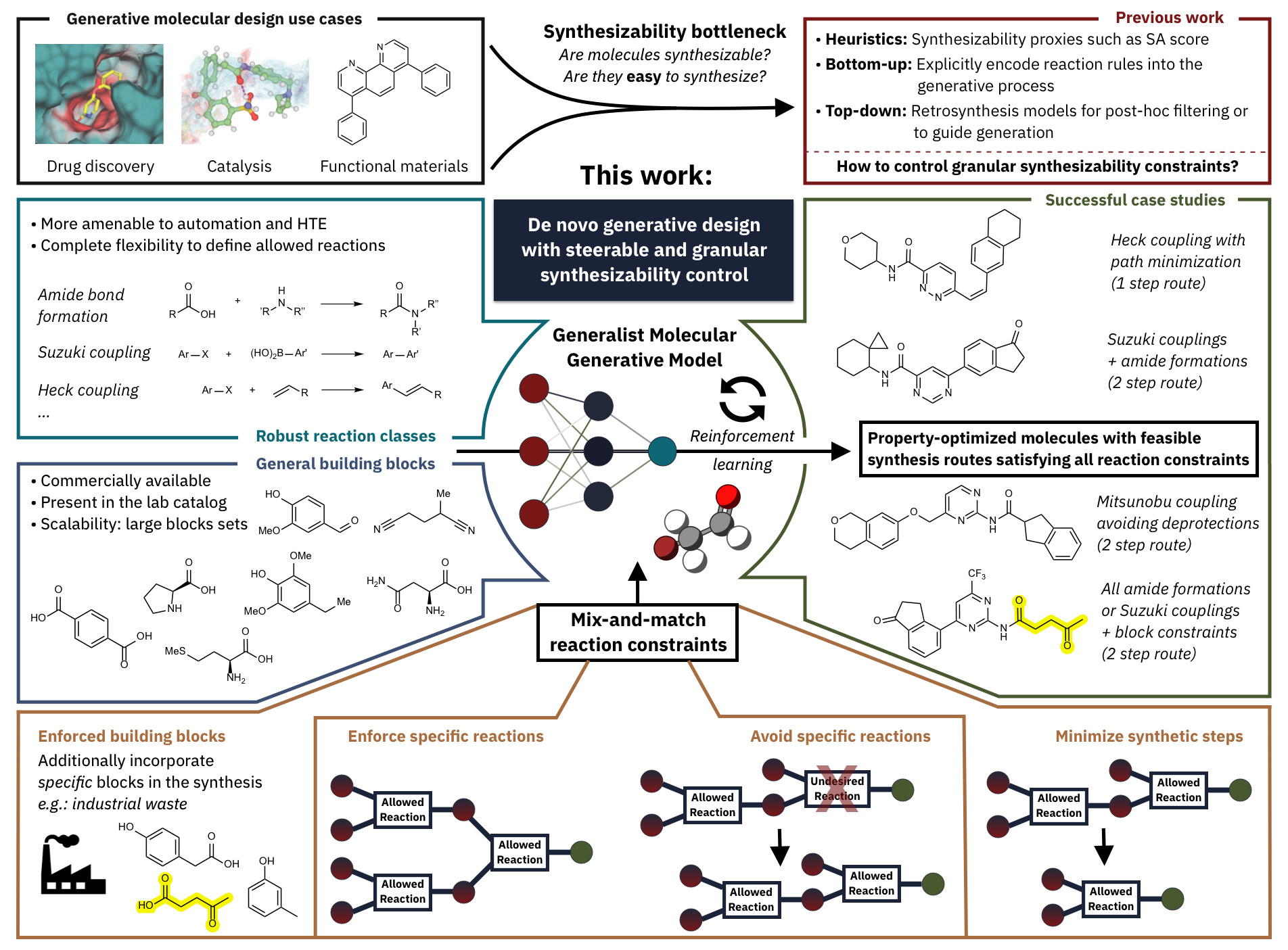
Generating Synthesizable Small Molecules
Generate small molecules predicted to be synthesizable using specific building blocks, specific reactions, avoiding specific reactions, and minimizing synthesis route length. Enable users to mix-and-match these synthesizability constraints. Generated molecules also satisfy multi-parameter optimization objectives (e.g. predicted binding affinity).
arXiv 2025
Generative Molecular Design with Steerable and Granular Synthesizability Control
J. Guo*, V. Sabanza-Gil*, Z. Jončev, J. S. Luterbacher, P. Schwaller
Generative molecular design with mix-and-match synthesizability constraints.
Nature Computational Science (Accepted) 2026
It Takes Two to Tango: Directly Optimizing for Constrained Synthesizability in Generative Molecular Design
J. Guo, P. Schwaller
Generate molecules with synthesis routes incorporating a small set of specific building blocks.
Chemical Science 2024
Directly Optimizing for Synthesizability in Generative Molecular Design using Retrosynthesis Models
J. Guo, P. Schwaller
Generate molecules with predicted synthesis routes.
Sample-efficient Optimization
Different drug discovery projects require molecules with different property profiles. A malleable generative model that can be tuned to satisfy arbitrary optimization objectives is practically useful. What aspects and components of the generative model and optimization algorithm can enhance sample efficiency (i.e. efficiently generate property-optimal molecules)?
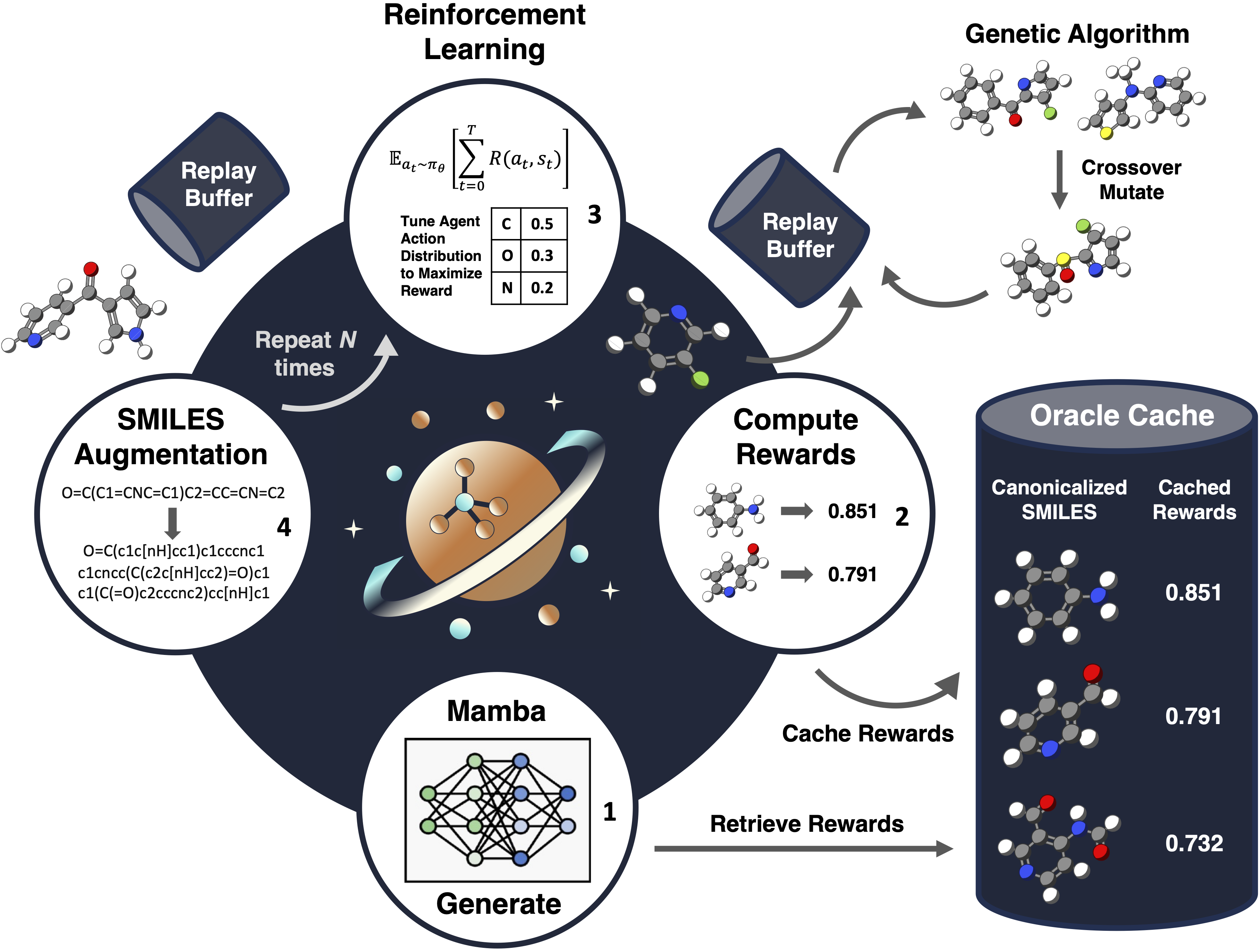
Nature Machine Intelligence (Accepted) 2026
Sample-efficient Generative Molecular Design using Memory Manipulation
J. Guo, J. Chen, A. GX-Chen, P. Schwaller
Framework (Saturn) for generative molecular design and understanding why and how experience replay with SMILES augmentation improves sample efficiency.
ICLR 2024
Beam Enumeration: Probabilistic Explainability For Sample Efficient Self-conditioned Molecular Design
J. Guo, P. Schwaller
Extracting molecular substructures from generative models can provide sample efficiency gains and a notion of explainability.
JACS Au 2024
Augmented Memory: Sample-Efficient Generative Molecular Design with Reinforcement Learning
J. Guo, P. Schwaller
Combining experience replay with SMILES augmentation can substantially improve sample efficiency.
Chemical Science 2024
Sample efficient reinforcement learning with active learning for molecular design
M. Dodds, J. Guo, T. Löhr, A. Tibo, O. Engkvist, J. P. Janet
Integrating active learning with reinforcement learning can improve sample efficiency.
Nature Machine Intelligence 2022
Improving de novo molecular design with curriculum learning
J. Guo*, V. Fialková*, J. D. Arango, C. Margreitter, J. P. Janet, K. Papadopoulos, O. Engkvist, A. Patronov
Decompose a complex optimization task into simpler, sequential tasks to accelerate convergence.
Education
École Polytechnique Fédérale de Lausanne (EPFL)
Ph.D. in Chemistry and Chemical Engineering
Advisor: Philippe Schwaller
Thesis: Language-based Sample-efficient and Synthesizable Molecular Generative Design
Imperial College London
MRes. Drug Discovery and Development
McGill University
B.Sc. Chemistry